More Examples¶
This page contains further examples of applications of Python scripts to Astronomy beyond those in the Quick tour of Python:
Reading a table and plotting with asciitable¶
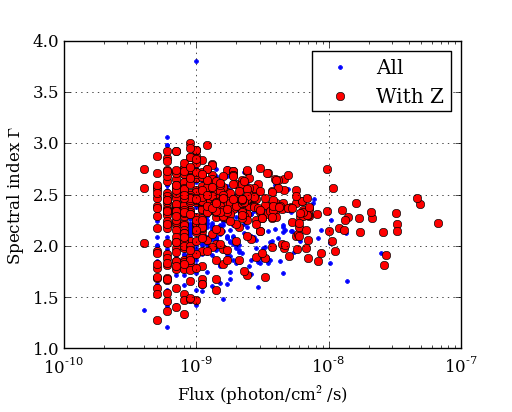
The Fermi Gamma-ray satellite has a nice catalog of AGN available through HEASARC. The script below will read in the catalog data using the asciitable module, do some basic filtering with NumPy, and make a couple of plots with matplotlib
import asciitable # Make external package available
# Read table.
# ==> dat[column_name] and dat[row_number] both valid <==
dat = asciitable.read('fermi_agn.dat')
redshift = dat['redshift'] # array of values from 'redshift' column
flux = dat['photon_flux']
gamma = dat['spectral_index']
# Select rows that have a measured redshift
with_z = (redshift != -999)
figure(1)
semilogx(flux, gamma, '.b', label='All') # First plot!
semilogx(flux[with_z], gamma[with_z], 'or', label='With Z')
legend(numpoints=1)
grid()
xlabel('Flux (photon/cm$^2$/s)') # latex works
ylabel('Spectral index $\Gamma$')
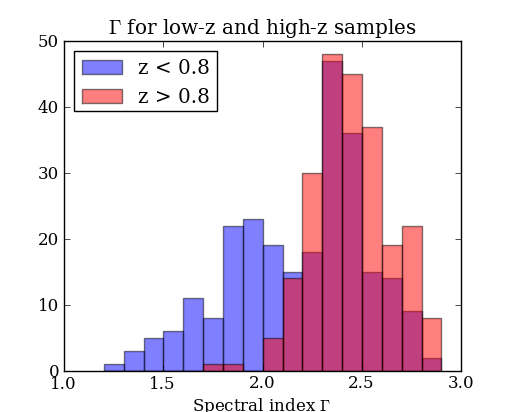
# Select low- and high-z samples
lowz = with_z & (redshift < 0.8)
highz = with_z & (redshift >= 0.8)
figure(2)
bins = arange(1.2, 3.0, 0.1) # values from 1.2 to 3.0 by 0.1
hist(gamma[lowz], bins, color='b', alpha=0.5, label='z < 0.8')
hist(gamma[highz], bins, color='r', alpha=0.5, label='z > 0.8')
xlabel('Spectral index $\Gamma$')
title('$\Gamma$ for low-z and high-z samples')
legend(loc='upper left')
asciitable.write(dat[with_z], 'fermi_agn_with_z.dat')
Curve fitting with SciPy¶
SciPy provides curve_fit, a simple and useful implementation of the Levenburg-Marquardt non-linear minimization algorithm. This example shows a code to generate a fake dataset and then fit with a gaussian, returning the covariance matrix for parameter uncertainties.
from scipy.optimize import curve_fit
# Create a function
# ==> First encounter with *whitespace* in Python <==
def gaussian(x, a, b, c):
val = a * exp(-(x - b)**2 / c**2)
return val
# Generate fake data.
# Note: functions in random package, array arithmetic (exp)
n = 100
x = random.uniform(-10., 10., n)
y = exp(-(x - 3.)**2 / 4) * 10. + random.normal(0., 2., n)
e = random.uniform(0.1, 1., n)
# Fit
popt, pcov = curve_fit(gaussian, x, y, sigma=e)
# Print results
print "Scale = %.3f +/- %.3f" % (popt[0], sqrt(pcov[0, 0]))
print "Offset = %.3f +/- %.3f" % (popt[1], sqrt(pcov[1, 1]))
print "Sigma = %.3f +/- %.3f" % (popt[2], sqrt(pcov[2, 2]))
# Plot data
errorbar(x, y, yerr=e, linewidth=1, color='black', fmt=None)
# Plot model
xm = linspace(-10., 10., 100) # 100 evenly spaced points
plot(xm, gaussian(xm, popt[0], popt[1], popt[2]))
# Save figure
savefig('fit.png')
The plotted fit result is as shown below:

Intermission: NumPy, Matplotlib, and SciPy¶
These three packages are the workhorses of scientific Python.
- NumPy is the fundamental package for scientific computing in Python [NumPy Reference]
- Matplotlib is one of many plotting packages. Started as a Matlab clone.
- SciPy is a collection of mathematical algorithms and convenience functions [SciPy Reference]
Synthetic images¶
This example demonstrates how to create a synthetic image of a cluster, including convolution with a Gaussian filter and the addition of noise.
import pyfits
from scipy.ndimage import gaussian_filter
# Create empty image
nx, ny = 512, 512
image = zeros((ny, nx))
# Set number of stars
n = 10000
# Generate random positions
r = random.random(n) * nx
theta = random.uniform(0., 2. * pi, n)
# Generate random fluxes
f = random.random(n) ** 2
# Compute position
x = nx / 2 + r * cos(theta)
y = ny / 2 + r * sin(theta)
# Add stars to image
# ==> First for loop and if statement <==
for i in range(n):
if x[i] >= 0 and x[i] < nx and y[i] >= 0 and y[i] < ny:
image[y[i], x[i]] += f[i]
# Convolve with a gaussian
image = gaussian_filter(image, 1)
# Add noise
image += random.normal(3., 0.01, image.shape)
# Write out to FITS image
pyfits.writeto('cluster.fits', image, clobber=True)
The simulated cluster image is below:

Running existing compiled codes¶
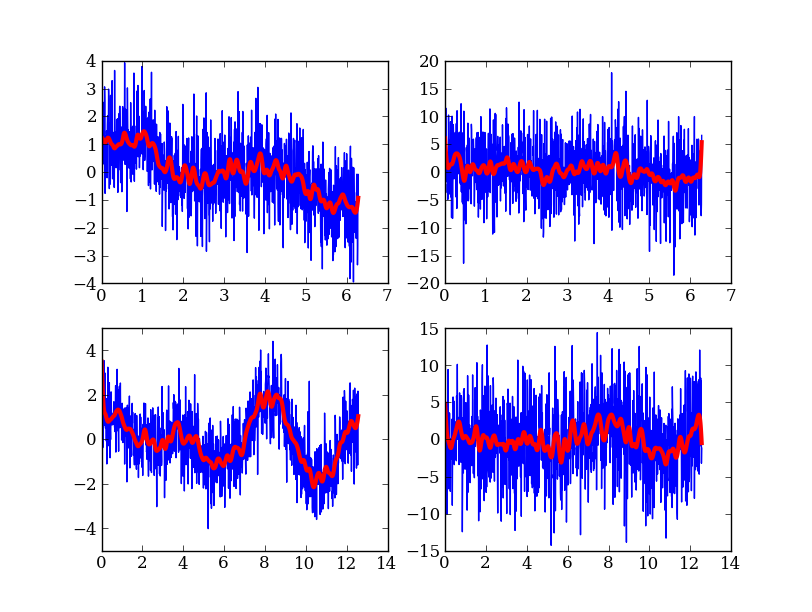
In addition to just doing computations and plotting, Python is great for gluing together other codes and doing system type tasks.
import os
import asciitable
smoothing = 30 # Smoothing window length
freqs = [2, 4] # Frequency values for making data
noises = [1, 5] # Noise amplitude inputs
figure(1)
clf()
# Loop over freq and noise values, running standalone code to create noisy data
# and smooth it. Get the data back into Python and plot.
plot_num = 1
for freq in freqs:
for noise in noises:
# Run the compiled code "make_data" to make data as a list of x, y, y_smooth
cmd = 'make_data %s %s %s' % (freq, noise, smoothing)
print 'Running', cmd
out = os.popen(cmd).read()
# out now contains the output from <cmd> as a single string
# Write the output to a file
filename = 'data_%s_%s' % (freq, noise)
open(filename, 'w').write(out)
# Parse the output string as a table
dat = asciitable.read(out)
# Make a plot
subplot(2, 2, plot_num)
plot(dat['x'], dat['y'])
plot(dat['x'], dat['y_smooth'], linewidth=3, color='r')
plot_num += 1
And much much more...¶
- Fast access to big (1e9 rows) tables with PyTables + HDF5
- 3-d plotting and surface rendering with Mayavi
- Sophisticated data modeling with advanced statistics with Sherpa
- Query VO tables and broadcast or retrieve tables to VO applications like TOPCAT.
- GUI application to quickly view thousands of X-ray survey image cutouts
- Python-based web site for browsing a complex multi-wavelength survey
- Thermal modeling of the Chandra X-ray satellite
- Interactive multi-user plots accessed through a web browser (!)
- Distributed computing with MPI for Python
- Make a little video distribution web site